Development News Brief
Get Galaxy
*** Upgrade NOW for an Important Security Fix ***
 |
getgalaxy.org | |
| new: | $ hg clone http://www.bx.psu.edu/hg/galaxy galaxy-dist |
|
| upgrade: | $ hg pull -u -r 12fcd068b12e |
|
# Communications and Collaboration
Administrators are always encouraged to install the latest release not only to incorporate new features, but to take advantage of the current set of security and bug fixes inherent in any project with a fast paced development cycle. This is even more strongly advised for those running public instances. The Galaxy team takes security very seriously and strives to quickly and clearly communicate when a security issue is identified. We have an amazing community that is instrumental in the discovery and resolution of problems when they are uncovered - and we want to thank them for their contributions and support. Galaxy is truly a team effort and transparency and information flow is our priority.
We also intend to learn from recent actions in these areas that fell short of a best effort. The security corrections and priority of the changes to the Compute tool in the prior distribution lacked the details and urgency notice it deserved. The move from Bitbucket to Trello for community collaboration in issue reporting also lacked the information and timely documentation that would have made the transition processes smoother. Both are covered in more detail in this News Brief. Our goals are to provide a remedy for the immediate issues and to create a successful communications and collaborations model for the future. We value your feedback - please let us know how we are doing as the new tools are put into practice.
# Compute Tool Security Fix
***This distribution and the previous distribution, October 5, 2012, fix a significant security hole in Galaxy's "Compute" tool. Upgrade or patch NOW. ***
The Compute columns tool allowed arbitrary execution of python code entered into the input text field by a user. It has been modified to only allow safe operators and functions. Administrators are strongly urged to apply this update for security purposes, or to manually apply/patch these changesets:
https://bitbucket.org/galaxy/galaxy-dist/changeset/b5bda7a5c34535ada63722941f2a2d62524b1faa
https://bitbucket.org/galaxy/galaxy-dist/changeset/685a17af92dfd6a2e3d7e3c9a3a4b119c78a6f96
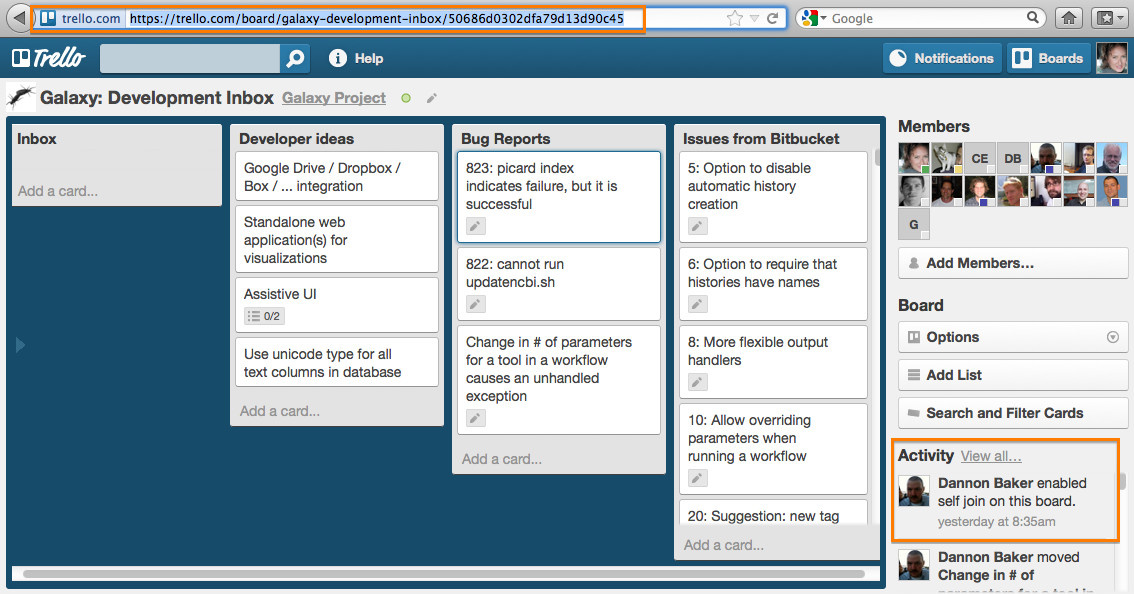
# New Trello Issue Board
The Galaxy project now has a new issue tracking system. The public facing Trello board is accessible from **http://galaxyproject.org/trello''' to perform simple web issue reporting. The direct link into the ''Trello'' board is '''https://trello.com/board/galaxy-development-inbox/50686d0302dfa79d13d90c45'''. Please note that the same guidelines about creating issues used previously at ''Bitbucket'' apply here as well - report software issues and change requests, but get support and ask questions on the mailing list: galaxy-dev@bx.psu.edu.
To add new cards, the community can quickly fill out the form at http://galaxyproject.org/trello and submit (use markdown for formatting, if desired), but comment on and vote for issues directly on the Trello issue board itself.
All existing galaxy-central bitbucket issues were imported and, though they still need to be sorted out, comments/voting on them are more than welcome. Our hope is that this issue tracker will be a more effective community interface than the Bitbucket issues were, with the voting features and simpler visibility.
The Galaxy wiki and other related resources have now been updated to point to the new tracking system. But if you happen to come across something we missed, please feel free to edit the wiki (if you are a vetted editor or decide to become one), or send us an email and we'll update.
# Tool Shed
Tool Shed Enhancements inside of Galaxy
- Enhance Galaxy's handling of Tool Shed Repositories to work with hierarchical relative paths.
- Allow directly going to a Tool Shed Repositories Manage page when you get the
already installed message. Makes it significantly easier to activate or reinstall an uninstalled/deactivated repository. - More graceful handling of uninstalling or deactivating toolshed repositories that have already been removed from disk.
- Enable activating and reinstalling Repositories directly from the manage repository page.
- Enhance Repository deactivate or uninstall page to better handle a repository when it has been been deactivated, but not uninstalled.
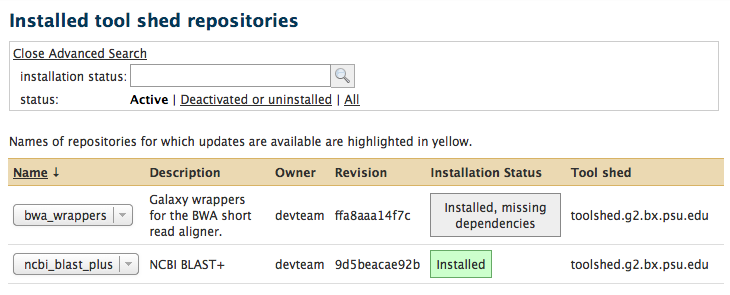
Galaxy enhancements for installed tool shed repositories
- Galaxy administrators can now reset metadata on installed tool shed repositories
We recommend that you reset the metadata on your installed tool shed repositories when you upgrade your Galaxy instance to this revision!
When a tool shed repository is installed into your local Galaxy instance, metadata is generated for the repository as a part of the installation process and stored in the tool_shed_repository.metadatatable column in the Galaxy database. This automatic process inspects the contents of the specific revision of the installed repository and generates and stores important information about it. This metadata information is used by certain Galaxy features. As new features are added to Galaxy or the tool shed, the process that generates this installed tool shed repository metadata within the Galaxy instance may be enhanced to accommodate information about the new features. Read more…
- #2 The Galaxy tool shed repository installation process has been enhanced to enable installation of repositories into a hierarchy of directories instead of a single level of directories.
If your Galaxy instance includes currently installed tool shed repositories, we recommend that you reset the metadata on your installed repositories as described in the above item, as doing this will reset certain information about your repositories, including entries for tools in your shed-related tool panel config files (e.g., shed_tool_conf.xml, migrated-tools_conf.cml, etc ).
Prior to this enhancement, values for the "tool_path" attribute defined in the <toolbox> tag within shed-related tool panel config files (e.g., shed_tool_conf.xml) were restricted to a single subdirectory level, something like this:
<toolbox tool_path="../shed_tools">
However, now these values can include subdirectory hierarchies like this:
<toolbox tool_path="../shed_tools/galaxy_instance1">
This allows the Galaxy administrator greater flexibility in defining locations within a single hierarchy for installed tool shed repositories using multiple shed-related tool panel config files. For example, a Galaxy administrator may now define installation locations like this.
tool panel config file name <toolbox> tag
--------------------------- -------------------
migrated_tools_conf.xml <toolbox tool_path="../shed_tools/galaxy_instance1/migrated_tools">
shed_tool_conf.xml <toolbox tool_path="../shed_tools/galaxy_instance1/installed_tools">
- #3 The process for handing uninstalling or deactivating tool shed repositories whose contents have previously been removed from disk has been streamlined by now displaying a message that includes a link that when clicked will handle the process.
- #4 When managing installed tool shed repositories, the previous "deleted" link in the Advanced Search feature has been enhanced to clarify that, when clicked, repositories that have been deactivated or uninstalled will be displayed.
Tools
- Add interpreter and absolute path logic to version tag; thanks to Björn Grüning.
- Tool search: remove Whoosh handling for Python version 2.4 and remove minscore.
- Performance enhancements for Build custom track for UCSC genome browser tool.
-
Add genomespace tools to
tool_conf.xml.main.- Get Data -> GenomeSpace import
- Send Data -> GenomeSpace export
- GenomeSpace now also enabled on Galaxy Main.
# Visualization framework
Trackster
- Fix: Show complete feature at bottom of track rather than cut it off.
Circster
- Can add datasets to visualization.
- Initiate and handle dataset indexing + dataset loading.
- Add visual indicator when loading data.
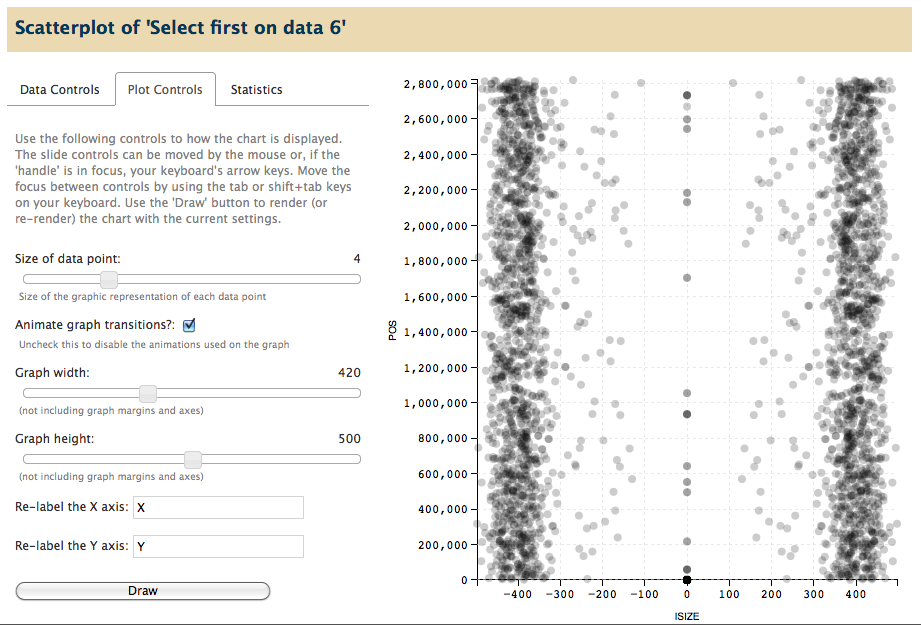
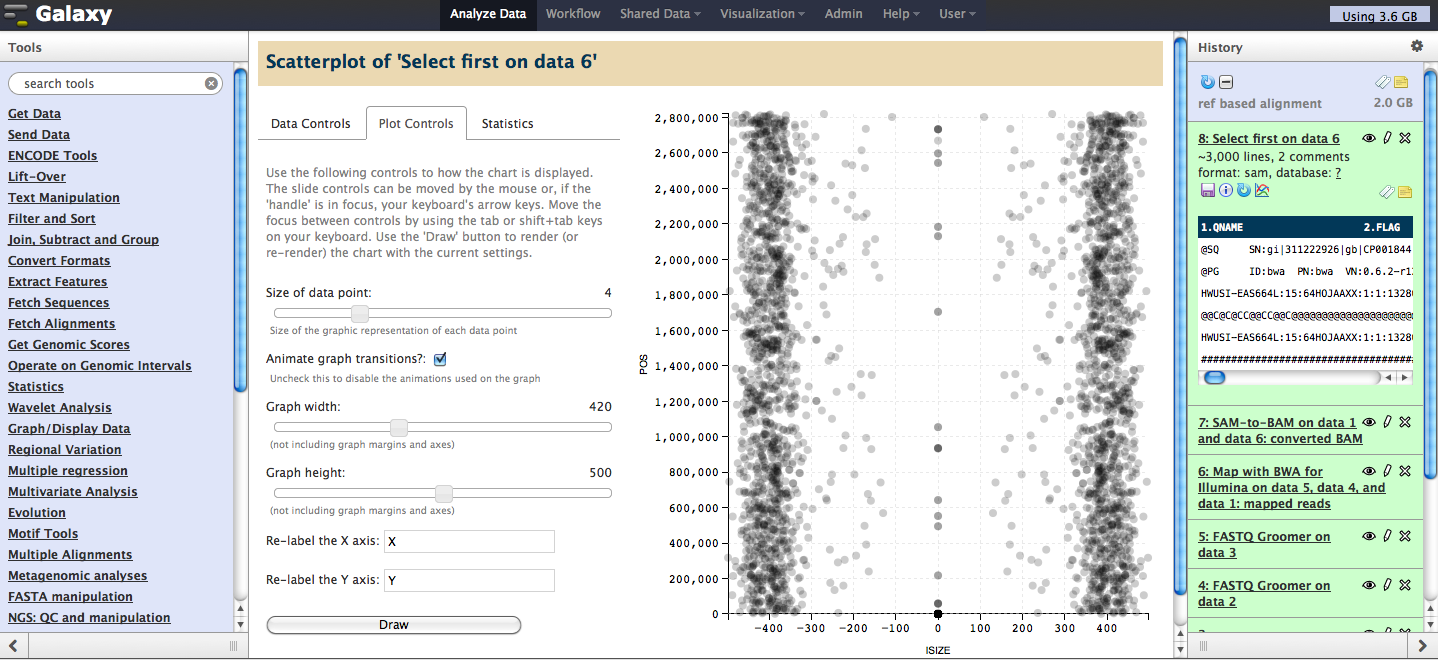
Scatter Plot
What it does: Given two numeric columns from a tabular dataset file, the scatter plot visualization function draws each datapoint onto a cartesian plane.
Quick start to graphing data:
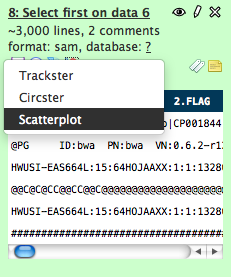
- Click on the visualizations icon of a tabular dataset.
- #2 Next, select 'Scatterplot' from the popup menu that appears and the main panel will load with data and graph options. Chose which two columns to load as datapoints and click the 'Draw' button to display the graph.
- #3 Tool is still undergoing further development and full documentation is pending, but here are the key details.
Basic specifications, functions, and features:
- Datapoints are drawn semi-transparent to convey density of multiple points.
- Axes are generated automatically based on the min/max of each numeric column.
- Grid lines are added in the background to facilitate locating the point on either access.
- Display the (x, y) of any specific point by hovering over it with the mouse (the given point is colored red and lines are drawn to each axis to assist). Please note: point detail display is unavailable in Chrome at this time.
- General statistics (count, min, max, mean, median) for each column are shown in a separate panel.
Additional user controlled functions:
- Set the columns from which the data are drawn.
- Add an optional third id column to associate with the other two columns. This id is then displayed, along with (x,y) values, when the user hovers over any given point. Please note: like point detail display, id display is unavailable in Chrome at this time.
- Control the dimensions of the graph, the size of datapoints, and the labels used for each axis.
Full screen view, showing scatter plot in the center panel:
API
- Enable dataset upload via tools controller; thanks to Brad Chapman.
- Enable querying of current history and user; thanks to Brad Chapman.
-
A new folders API has been developed. It permits operations in formats similar to the library API. Wiki documentation is pending, but for now, the available actions are defined as:
GET /api/folders/{FolderId}: This returns information about the folder.POST /api/folders/{FolderId}: Create a folder. The same parameters asPOST /api/libraries/{LibraryId}/contents/{FolderId}are utilized, except that thecreate_type parameteris ignored.GET /api/folders/{FolderId}/contents: This returns the folder's contents.
Source
- Allow admins to access datasets/jobs with access restrictions.
- Allow Admins to rerun jobs with access restrictions.
- Unhide failed output datasets.
- Do not hide failed datasets with
HideDatasetActionpost job action. - Library browsing optimizations. Opening libraries with many datasets and/or folders is now quicker.
Bug Fixes
-
Galaxy
- Fix for preview display of tabular items when certain metadata was not set.
- Do not use
backrefsin mapping forcopied_from_library_dataset/history _dataset_associationas it confuses SQLAlchemy (resulted in 'is not available, due to conflicting property'). - Correctly determine the inheritance and creating job for a dataset. Fixes issues with showing dataset info, rerun, viewing and reporting dataset errors.
- Fix for managing user info (e.g changing email/password) that appeared in last distribution.
- Fix bug in stoping user jobs in the admin interface.
- Custom
BioStarlinkouts (not fully implemented) are now off by default. - Some fixes for handling unicode data in the UI.
- Fix for dataset display.
- Various fixes for Python 2.5.
- Bugs introduced during the controller architecture migration (often resulting in error message such as "No module named controllers.XYZ") have been traced and resolved.
-
Tool Shed
- Fix for tool shed
get_category_by_namewhen category does not exist. - Better handle missing 'tool' entry in tool repository metadata.
- Fix for tool shed
Announcements
News, October 2012 Galaxy Update
# About Galaxy
The Galaxy Team is a part of BX at Penn State, and the Biology and Mathematics and Computer Science departments at Emory University.
Galaxy is supported in part by NSF, NHGRI, the Huck Institutes of the Life Sciences, and The Institute for CyberScience at Penn State, and Emory University.
Join us at Twitter @galaxyproject or just read our tweets Galaxy on Twitter