April 2016 Galaxy News
Welcome to the April 2016 Galactic News, a summary of what is going on in the Galaxy community.
If you have anything to include in the next News, please send it to [Galaxy Outreach](mailto:outreach AT galaxyproject DOT org).
GCC2016
GCC2016 will be held June 25-29 at Indiana University in Bloomington, Indiana, United States. This will be the 7th annual gathering of the Galaxy community, and we are expecting over 200 participants again this year. The 2016 Galaxy Community Conference includes 2 days of hackathons, 2 days of training, and a two day meeting featuring accepted presentations, keynotes, poster sessions, the new Visualization Showcase and Software Demo sessions, lightning talks, birds-of-a-feather meetups, and plenty of networking.
GCC2016 Abstract Deadline extended to April 8!
The deadline for oral presentation abstracts has been extended to April 8, 2016. Abstracts will be reviewed and submitters will be notified of acceptance status no later than April 22, 2016.
Poster and Demo abstract submission closes May 20, 2016 (or earlier depending on poster space availability). Poster and demo submissions are reviewed on a rolling basis, and submitters will be notified of acceptance status no later than two weeks after the abstract is submitted. You may submit similar content for oral presentations, posters, and demos.
Topics should be of interest to those working in high-throughput data analysis and research. Presentations that are Galaxy-centric are encouraged, but not required. Please see the abstracts page for full details.
GCC2016 Early Registration
Early registration for GCC2016 is now open. Registration costs depend on which events you register for, your career stage & affiliation, and when you register. Early bird registration ends May 20 and is up to 40% less than regular registration rates. Early bird registration starts at less than $45 / day for students and postdocs, and at $65 / day for other attendees from non-profits.
You can also sign up for conference housing during registration.
You are strongly encouraged to review the training and housing options before beginning the registration process.
Scholarships: Application deadline is May 1
We are pleased to offer scholarships for the 2016 Galaxy Community Conference, being held in Bloomington, Indiana, United States, June 25-29. Scholarships are available to students and post-docs in historically under-represented groups, and to those from or based in Low and Lower-Middle Income Economies, as defined by the World Bank. If this describes you or one of your students then we hope to receive an application.
Scholarships cover registration and lodging during the GCC Meeting, and for any Training or Hackathon events the applicant chooses to attend. Scholarships do not cover travel or other expenses. The application deadline is May 1 for members of historically underrepresented groups.
See the full announcement for details.
Sponsors
Please welcome the Indiana Health Industry Forum (IHIF) as a new GCC2016 Sponsor. IHIF is a statewide trade association representing members of Indiana's health science business community. Our mission is to connect key stakeholders to: enhance business networks, advocate for member interests, develop workforce skills, and provide strategic vision in the interest of growing the state’s health industry economy and reputation.
We continue to seek other sponsors as well and offer a wide range of sponsorship plans. If your organization is interested in having a presence at GCC2016, please contact the GCC2016 Exec for more information.
Events
Galaxy DevOps Workshop - Heidelberg, 6-7 April
We would like to invite you to our Galaxy DevOps Workshop in Heidelberg from 6-7 of April, organised by de.NBI and ELIXIR.
The workshop is intended for Bioinformaticians who want to
- learn the internals of the multi-omics Galaxy workbench
- learn how to set up and run a Galaxy server
- learn about Galaxy tool development
- get questions answered from experienced Galaxy admins and community members
- are interested in reproducible and accessible research
The workshop will be held at Heidelberg Center for Human Bioinformatics (HD-HuB). Location and Accommodation options can be found here.
Registration is open and their is still space - just send Björn Grüning an email. For more information please see the workshop page.
Hackathon: Tools, Workflows and Workbenches, 18-20 May
A hackathon bringing together developers from the ELIXIR Tools & Data Services Registry, Galaxy, Taverna, Arvados, CWL, ReGaTE and EDAM ontology, with Galaxy instance providers from ELIXIR and beyond, to promote collaboration and technical developments will take place on 18-20 May 2016 at the Institut Pasteur in Paris.
Registration is free but space is limited.
Report: IUC Contribution Fest - RADSeq Tools and Workflows
A remote Contribution Fest was held 7th and 8th of March for developers to work on Galaxy RADSeq tools. RADSeq is a cheap sequencing technology that is used by many resource-limited groups who would benefit a lot from easy-to-use galaxy tools. Indeed there has been quite some interest in analyzing RADSeq with Galaxy. Currently there is a wrapper for stacks and little more to help with RAD specific analysis (though many other galaxy tools are useful with RADs - bwa, cap3, gatk, velvet, ...).
A hackathon report page is available on the GenOuest GUGGO wiki.
Upcoming Events
There are many upcoming events in the next few months. See the Galaxy Events Google Calendar for details on other events of interest to the community.
| |
Designates a training event offered by GTN member(s) |
New Papers
86 new papers referencing, using, extending, and implementing Galaxy were added to the Galaxy CiteULike Group in March.
It was a banner month for Galaxy-related papers. Some of the highlights:
- The Language Application Grid Nancy Ide, James Pustejovsky, Christopher Cieri, et al., edited by Yohei Murakami, Donghui Lin. In Worldwide Language Service Infrastructure, Vol. 9442 (2016), pp. 51-70, doi:10.1007/978-3-319-31468-6_4
- NGS-QC Generator: A Quality Control System for ChIP-Seq and Related Deep Sequencing-Generated Datasets Marco Antonio Mendoza-Parra, Mohamed-AshickM Saleem, Matthias Blum, Pierre-Etienne Cholley, Hinrich Gronemeyer, edited by Ewy Mathé, Sean Davis. In Statistical Genomics, Vol. 1418 (2016), pp. 243-265, doi:10.1007/978-1-4939-3578-9_13
- Galaxy-M: a Galaxy workflow for processing and analyzing direct infusion and liquid chromatography mass spectrometry-based metabolomics data Robert L. Davidson, Ralf J. M. Weber, Haoyu Liu, Archana Sharma-Oates, Mark R. Viant. GigaScience, Vol. 5, No. 1. (23 February 2016), doi:10.1186/s13742-016-0115-8
- Models and Simulations as a Service: Exploring the Use of Galaxy for Delivering Computational Models Mark A. Walker, Ravi Madduri, Alex Rodriguez, Joseph L. Greenstein, Raimond L. Winslow. Biophysical Journal, Vol. 110, No. 5. (March 2016), pp. 1038-1043, doi:10.1016/j.bpj.2015.12.041
- ZBIT Bioinformatics Toolbox: A Web-Platform for Systems Biology and Expression Data Analysis Michael Römer, Johannes Eichner, Andreas Dräger, Clemens Wrzodek, Finja Wrzodek, Andreas Zell. PLoS ONE, Vol. 11, No. 2. (16 February 2016), e0149263, doi:10.1371/journal.pone.0149263
- A User-Friendly Computational Workflow for the Analysis of MicroRNA Deep Sequencing Data Anna Majer, KyleA Caligiuri, StephanieA Booth. In MicroRNA Protocols, Vol. 936 (2013), pp. 35-45, doi:10.1007/978-1-62703-083-0_3
The new papers were tagged with:
| # | Tag | # | Tag | # | Tag | # | Tag | |||
|---|---|---|---|---|---|---|---|---|---|---|
| - | Cloud | 6 | Other | 3 | Shared | 14 | UseMain | |||
| 3 | HowTo | - | Project | 5 | Tools | 17 | UsePublic | |||
| 5 | IsGalaxy | 9 | RefPublic | - | UseCloud | - | Visualization | |||
| 44 | Methods | 4 | Reproducibility | 4 | UseLocal | 17 | Workbench |
Who's Hiring

The Galaxy is expanding! Please help it grow.
- Research Associate, University of Bristol Elizabeth Blackwell Institute and Public Health England, Bristol, United Kingdom
- Postdoctoral researcher, University of Freiburg, Freiburg, Germany.
- Galaxy Tool Development for small RNA biology, epigenetics & viruses metagenomics, Institut de Biologie Paris Seine, Paris, France
- Intégration d'outils pour la métagénomique phytovirale dans Galaxy, INRA, Toulouse, France
- Core Computational Biologist / Bioinformatician, The Sainsbury Lab, Norwich, United Kingdom. Closes 15 April
- Bioinformatics Web Application Developer-Biology (Job ID 32899), Washington University in St. Louis, Missouri, United States
- Software developer and Post-docs, Gehlenborg Lab, Harvard Medical School, Boston, Massachusetts, United States
- Postdoctoral Research Positions, Molecular and Cellular Biology Department at Baylor College of Medicine, Houston, Texas, United States
- Bioinformaticist, Johns Hopkins University Applied Physics Laboratory (APL)
- Software Engineer, Oregon Health Sciences University, Portland, Oregon, United States
Got a Galaxy-related opening? Send it to outreach@galaxyproject.org and we'll put it in the Galaxy News feed and include it in next month's update.
New Public Galaxy Servers
1 new publicly accessible Galaxy server was listed in March:
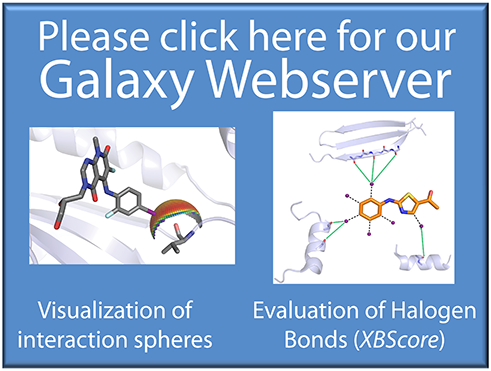
Halogen Bonding
-
Link:
-
Domain/Purpose:
- Visualize halogen bond contacts in the protein binding site
- Analyze a protein binding site for halogen bonding hotspots
- Evaluate halogen contacts with the protein backbone using the scoring function XBScore
-
Comments:
-
Supports visualizations (halogen bond spheres) for:
- the protein backbone (spherical scans and planar scans)
- methionine
- histidine
- More visualizations and evaluation tools will be added in the near future.
-
See
- Evaluating the Potential of Halogen Bonding in Molecular Design: Automated Scaffold Decoration Using the New Scoring Function XBScore, Markus O. Zimmermann, Andreas Lange, and Frank M. Boeckler Journal of Chemical Information and Modeling 2015 55 (3), 687-699 DOI:10.1021\ci5007118
- Using halogen bonds to address the protein backbone: a systematic evaluation, Rainer Wilcken, Markus O. Zimmermann, Andreas Lange, Stefan Zahn, and Frank M. Boeckler J. Comput. Aided Mol. Des. 2012, 26 (8), 935-945. DOI:10.1007/s10822-012-9592-8
-
- User Support:
-
Quotas:
- "Files older than 30 days will be deleted, so make sure to save the results to your hard drive. "
-
Sponsor(s):
- This webserver is provided by the group of Prof. Dr. Frank Boeckler (University of Tuebingen, Germany)
Galaxy Community Hubs



Share your training resources and experience now Share your experience now
Two new Community Log entries were added in March
- Testing connexion between Galaxy and iRODS by Mikael Loaec and Olivier Inizan
- Setting up Galaxy with TORQUE on AWS by Alex Kanterakis
As were two and two new training organizations.
- EMBL-ABR is a distributed national research infrastructure providing bioinformatics support to life science researchers in Australia.
- GenoToul bioinformatics platform, Sigenae, NED (GenPhySE) and TWB offers a catalog of training sessions on Galaxy.
Releases
Planemo 0.24.0
Planemo is a set of command-line utilities to assist in building tools for the Galaxy project. Planemo 0.24.0, the most recent release, features these updates:
- Drop support for Python 2.6. 93b7bda
- A variety of fixes for
shed_update. Pull Request 428, Issue 416 - Fix reporting of metadata updates for invalid shed updates. Pull Request 426, Issue 420
- Check
.shed.ymlowner against credentials during shed creation. Pull Request 425, Issue 246 - Fix logic error if there is a problem with
shed_create. 358a42c - Tool documentation improvements. 0298510, a58a3b8
See the release history.
CloudMan 16.03
We just released an update to Galaxy CloudMan on AWS. CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration or imposed quotas. Once running, you have complete control over Galaxy, including the ability to install new tools.
Most notable changes include:
- Galaxy 16.01 release
- A fine-grained control over auto-scaling options
- Several fixes to cluster sharing and cloning
See the CHANGELOG for a more complete set of changes.
Earlier Releases
Galaxy v16.01
The January 2016 (v16.01) release of Galaxy features
- Interactive Tours
- Wheels
- Nested Workflows
See the announcement for full details.
Galaxy administrators should also be aware of [these security announcements](/galaxy-updates/2016-04/#security-announcements) below. * [Read/write arbitrary filesystem paths, arbitrary code execution](https://lists.galaxyproject.org/archives/list/galaxy-dev@lists.galaxyproject.org/message/TXP6BGNZG5Y75ASM5HPQALI2UOPKXXTH/) * [Tool Shed Security Vulnerability - Read/write arbitrary filesystem paths](https://lists.galaxyproject.org/archives/list/galaxy-dev@lists.galaxyproject.org/message/TQG5K6DHSJX6HDFMV6MJ43SRNEO3X3JZ/)Galaxy Docker Image 16.01
And, thanks to Björn Grüning, there is also now a Docker image for Galaxy 16.01 as well.
galaxy-lib 16.4.0
galaxy-lib is a subset of the Galaxy core code base designed to be used as a library. This subset has minimal dependencies and should be Python 3 compatible. It's available from GitHub and PyPi.
CloudBridge 0.1.0
The Galaxy Team is proud to be part of the development team for a new cross-cloud library called CloudBridge. CloudBridge is a Python library providing a simple layer of abstraction over different cloud providers, reducing or eliminating the need to write conditional code for each cloud. The library is generally applicable to any domain wishing to run cloud-independent applications. There is already support for Amazon and OpenStack clouds with support for Google’s Compute Engine in development.
The first version of CloudBridge was released earlier this month and it comes with detailed user documentation. The source code is available on Github.
Starforge 0.1
Starforge is a collection of scripts that supports the building of components for Galaxy. Specifically, with Starforge you can:
- Build Galaxy Tool Shed dependencies
- Build Python Wheels (e.g. for the Galaxy Wheels Server)
- Rebuild Debian or Ubuntu source packages (for modifications)
These things will be built in Docker. Additionally, wheels can be built in QEMU/KVM virtualized systems.
Documentation can be found at starforge.readthedocs.org.
Pulsar 0.6.1
Pulsar 0.6 was released in December. Pulsar is a Python server application that allows a Galaxy server to run jobs on remote systems (including Windows) without requiring a shared mounted file systems. Unlike traditional Galaxy job runners - input files, scripts, and config files may be transferred to the remote system, the job is executed, and the results are transferred back to the Galaxy server - eliminating the need for a shared file system.
The 0.6.x release includes these changes:
- Pulsar now depends on the new galaxy-lib Python package instead of manually synchronizing Python files across Pulsar and Galaxy.
- Numerous build and testing improvements.
- Fixed a documentation bug in the code (thanks to @erasche). e8814ae
- Remove galaxy.eggs stuff from Pulsar client (thanks to @natefoo). 00197f2
- Add new logo to README (thanks to @martenson). abbba40
- Implement an optional awknowledgement system on top of the message queue system (thanks to @natefoo). Pull Request 82 431088c
- Documentation fixes thanks to @remimarenco. Pull Request 78, Pull Request 80
- Fix project script bug introduced this cycle (thanks to @nsoranzo). 140a069
- Fix config.py on Windows (thanks to @ssorgatem). Pull Request 84
- Add a job manager for XSEDE jobs (thanks to @natefoo). 1017bc5
- Fix pip dependency installation (thanks to @afgane) Pull Request 73
BioBlend 0.7.0
BioBlend version 0.7.0 was released at the beginning of November. BioBlend is a python library for interacting with CloudMan and the Galaxy API. CloudMan offers an easy way to get a personal and completely functional instance of Galaxy in the cloud in just a few minutes, without any manual configuration.) From the release CHANGELOG.
blend4j v0.1.2 blend4j v0.1.2 was released in December 2014. blend4j is a JVM partial reimplemenation of the Python library bioblend for interacting with Galaxy, CloudMan, and BioCloudCentral.
ToolShed Contributions
See list of tools contributed in March.
Automatic Testing of IUC Tools with Conda
An effort is underway to test more and more IUC tools automatically using Conda dependencies. The final aim of getting all tools under test by simply installing the dependencies via conda before running
planemo test. Getting packages in Conda also means we can automatically build Docker containers and offer Docker and Conda dependency resolutions for everyone today and for testing.The IUC is seeking comments and contributions to this effort.
Other News
- New and recently updated tools are now highlighted on usegalaxy.org.
-
From Bjöern Grüening:
- Cheminformatics datatypes were added to Galaxy with this pull request. Expect new ChemicalToolBox development soon!
- deepTools visualization, QC, and data normalization tools are now available on usegalaxy.org. Thanks to Devon Ryan and https://twitter.com/fidel_ramirez|Fidel Ramirez]]
- Galaxy Tours, a new interactive way to learn Galaxy, were announced in the February News. We now have an official repository to collect community tours.
-
From Yvan Le Bras:
- My first Galaxy Interactive Tour attempt for RADseq data analysis based on a RNAseq, using Björn Grüning's template.